Published in 2025
Deepflare achieves 100% success in vitro protein expression. Is it a new level of vaccine design?
In a recent collaboration with the leading French Biotech company Osivax, Deepflare demonstrated that for the most challenging viral vaccines, even Nobel-winning AI models like RFdiffusion can be of no use. Deepflare AI achieved a 100% expression rate (as demonstrated in a prospective in vitro test by Osivax), while RFdiffusion yielded 0% in silico success rate.
This result seemed almost too good to be true. So we asked ourselves - how can Deepflare’s models compare to RDdiffusion across bigger datasets? Our research team proceeded to develop a benchmark by including the dataset of the hardest antigens obtained from the original publication of David Baker, Nobel laureate behind RFdiffusion.
To compare two methods, we conducted a head-to-head comparison on two benchmarks. First, the epitope we were scaffolding for Osivax. Second, for the hardest B-cell epitopes from David Baker's original benchmark [2]
For each epitope, we generated thousands of sequences. Then, to measure the success rate, we computed the fraction of structures that closely resemble the original epitope. Success was defined according to David Baker's methodology, proven to highly correlate with experimental results [1]. redicted structures should have high confidence and extremely high geometric similarity in the epitope region: quantified by metrics - PAE<5, RMSD <2Å, and motif RMSD <1Å[2].
For the Osivax benchmark, RFdiffusion failed completely, even though we tested over 50000 different sequences; none of them passed through rigorous criteria. The Deepflare platform was able to generate tens of different structures that passed through the criteria, despite generating fewer than a thousand candidates. The ten best selected candidates were tested, and all of them could be expressed in E.Coli and then purified. For the simpler epitopes, in-silico methods achieved at best a 66% expression rate, after months of additional optimization.
Fig 1. % of successfully designed candidates expressed in E.coli and confirmed by the Western-Blot. Market standard estimation is based on experimental data of “RFjoint inpainting”[4]
Enabling surgical precision vaccines
To understand the significance of that technology, you need to imagine inducing neutralizing antibodies specific to only the most important B-cell epitope, without distracting the immune system with irrelevant, unstable, non-protective B-cell epitopes. This challenge requires designing a protein consisting of only a specific B-cell epitope conformation. In the same way as subunit vaccines did with the sequence. So let’s take those generalised results based on your benchmark to the specific case in particular, in the context of the hardest targets possible.
Case study: Precision response against RSV-site 0
RSV epitope site-0 is a great example of this design challenge. This epitope is crucial for neutralizing the virus, but it only appears before the virus fuses with a cell (pre-fusion). Standard vaccination with the full antigen is ineffective in this instance because the immune system mostly sees a changed (post-fusion) version of the epitope. Even with a stabilized pre-fusion antigen, the immune response is weakened because the immune system is distracted; antibodies are made against many non-neutralizing parts, and not just the epitope site-0.[3]
Fig 3a. The RSV epitope site-0, crucial for vaccine efficacy, is only a small part of the whole protein. After immunization, the antibody response will be diluted against multiple different epitopes.
Fig 3b. The designed antigen consists only of that epitope, enabling a highly specific immune response.
Why is Designing These Epitopes So Complex?
These crucial epitopes frequently consist of both well-defined secondary structures and highly flexible loops. A common problem is that in the generated designs by RFdiffusion, flexible loops incorrectly fold into secondary structures, disrupting the epitope's natural shape and shielding its antigenicity, minimizing vaccine efficacy.
Fig 4. Current de novo design tools like RFdiffusion struggle with this complexity, often generating designs with low confidence in the critical regions. Deepflare's platform resolves that problem, achieving higher metrics on the RFdiffusion authors' benchmark [1].
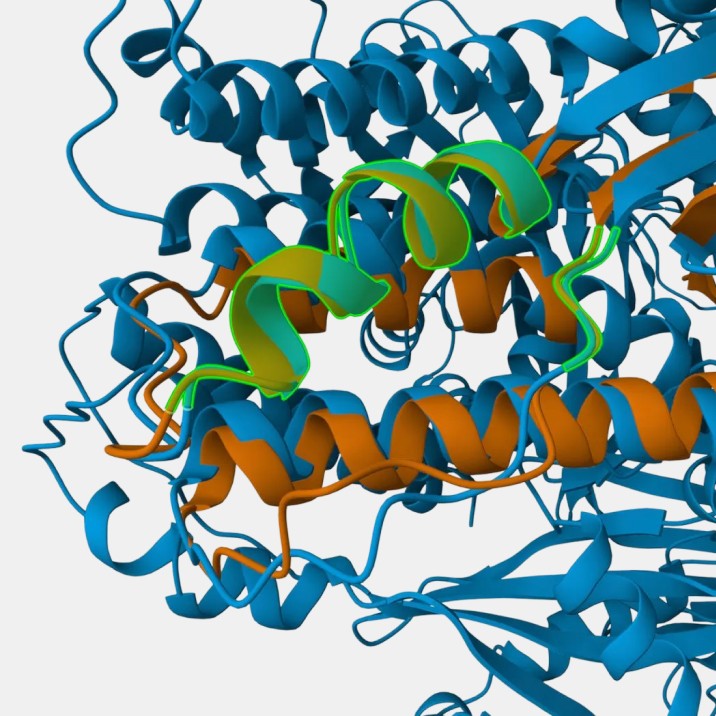
Fig 5a. Structural alignment of our candidate The highlighted parts are residues that bind to neutralizing antibodies.
Fig 5b. Structural alignment of the best RFdiffusion candidate.
Bibliography:
[1] Watson, J.L., Juergens, D., Bennett, N.R., Trippe, B.L., Yim, J., Eisenach, H.E., Ahern, W., Borst, A.J., Ragotte, R.J., Milles, L.F., et al. (2023). De novo design of protein structure and function with RFdiffusion. Nature, 620, 1089–1100.
[2] Watson, J.L., Juergens, D., Bennett, N.R., Trippe, B.L., Yim, J., Eisenach, H.E., Ahern, W., Borst, A.J., Ragotte, R.J., Milles, L.F., et al. (2023). Supplementary Information: De novo design of protein structure and function with RFdiffusion. Nature, 620
[3] Krarup, A., Truan, D., Furmanova-Hollenstein, P., Bogaart, G., Boks, L., Benkarim, P., Brouwer, I.J.M., Weterings, M.N., Zeldin, R., Melgert, B.N., et al. (2015). A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nature Communications, 6, 8143.
[4] Wang, J., Lisanza, S., Juergens, D., Tischer, D., Watson, J.L., Castro, K.M., Ragotte, R., Saragovi, A., Milles, L.F., Baek, M., et al. (2022). Scaffolding protein functional sites using deep learning. Science, 377(6604), 387–394.